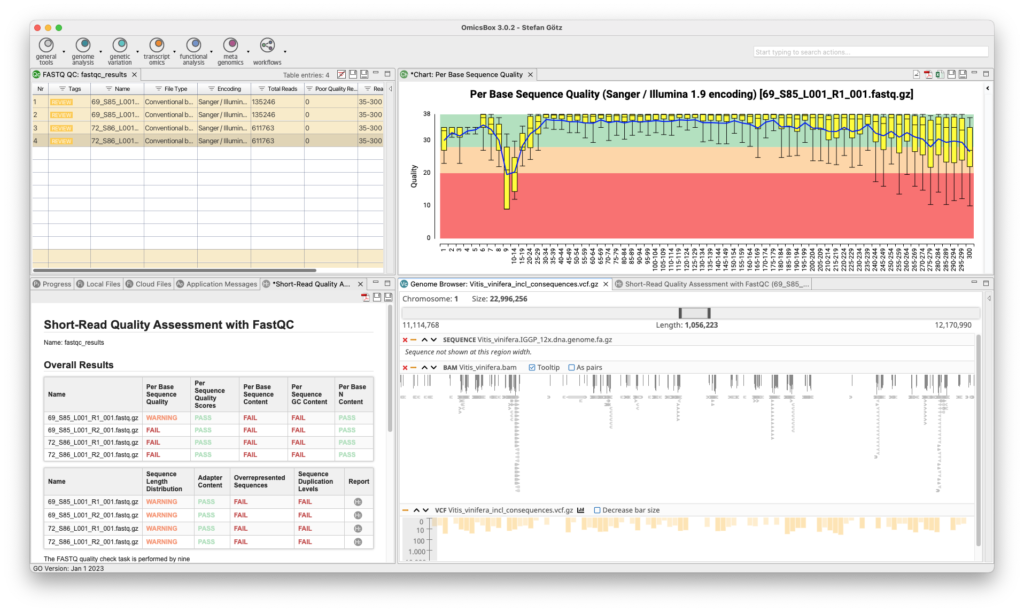
- Quality Control And Assessment
- De-Novo Assembly
- Repeat Masking
- Gene Finding
- Coding Potential
- Multi-locus Sequence Typing (MLST)
- Fast Variant Calling
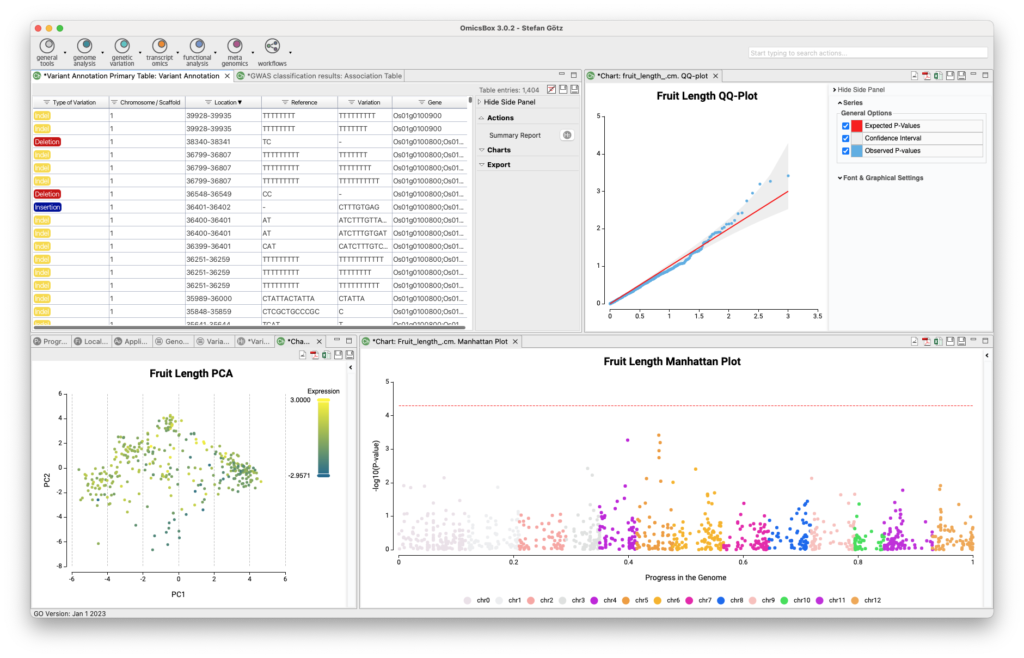
- Model & Non-Model Variant Annotation
- Variant Filtering
- Guided Genome-Wide Association Studies
- Supports GBS and WGS data
- Quality Control
- Quantify Expression
- De-Novo Assembly
- RNA-Seq Alignment
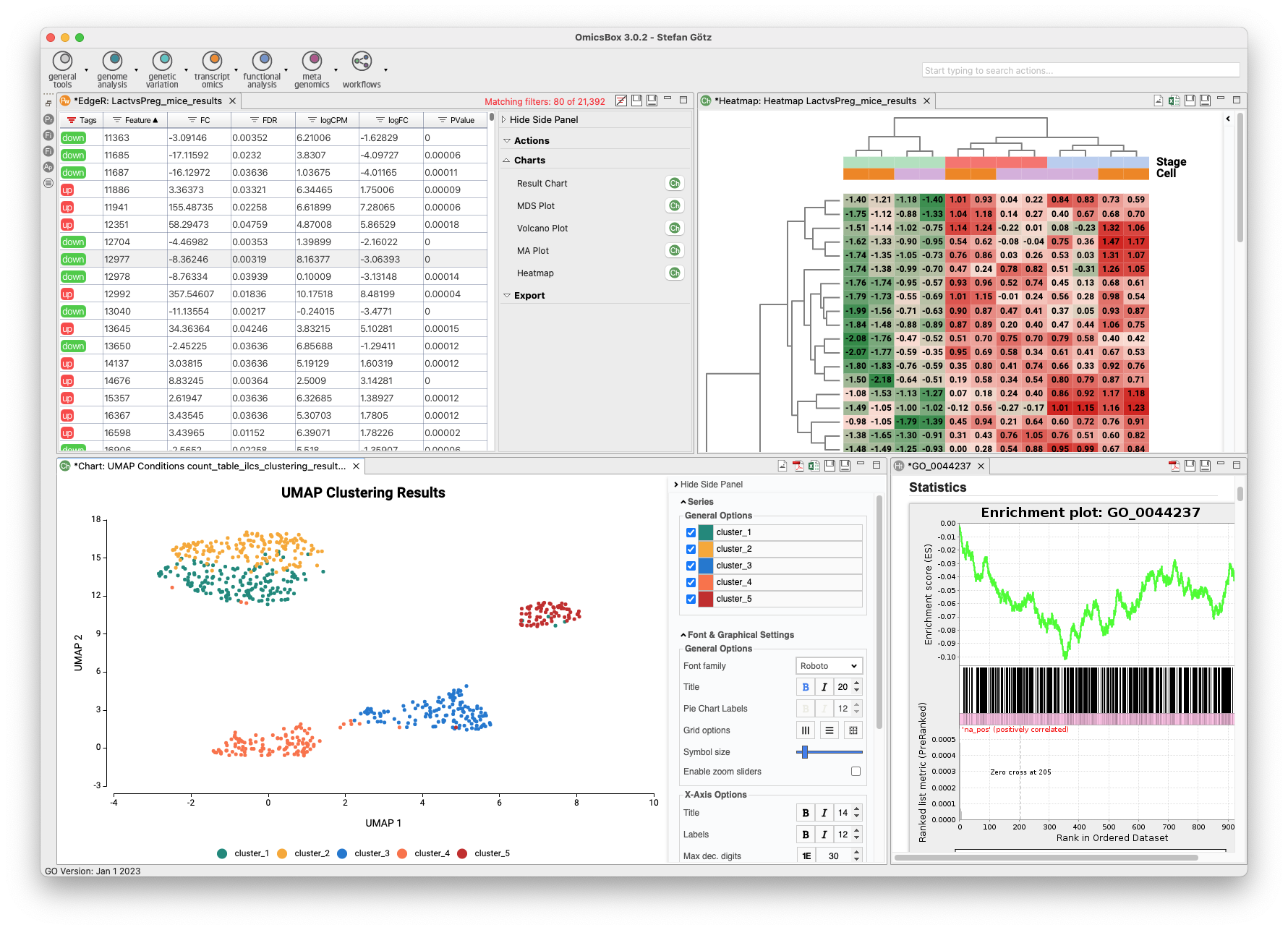
- Differential Expression Analysis
- Long-Read Analysis
- Single-Cell RNA-Seq
- High-Throughput Blast and InterProScan
- Gene Ontology Mapping
- Blast2GO Annotation
- Enrichment Analysis
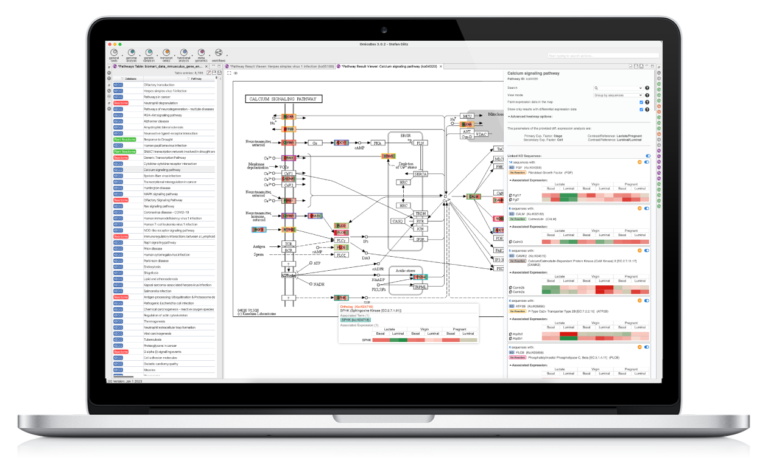
- Functional Interpretation
- Combined Pathway Analysis
- Quality Control And Assessment
- Taxonomic Classification
- Metagenomic Assembly
- Gene Prediction
- Functional Annotation
The All-In-One Bioinformatics Software
OmicsBox is a leading bioinformatics software that offers end-to-end NGS data analysis of genomes, transcriptomes, and metagenomes. The application is used by top private and public research institutions worldwide and allows researchers to easily process large and complex data sets, and streamline their analysis process. It is designed to be user-friendly, efficient, and with a powerful set of tools to extract biological insights from omics data.

User-Friendly

Versatile

Ready to use
OmicsBox Modules
The software is structured in different modules, each with a specific set of tools and functions designed to perform different types of analysis, such as de-novo genome assemblies, genetic variation analysis, differential expression analysis, and taxonomic classifications of microbiome data, including the functional interpretation and rich visualizations of results. The functional analysis module, which includes the popular Blast2GO annotation methodology makes OmicsBox particularly suited for non-model organism research. This is demonstrated by over 15k scientific research citations. OmicsBox works out of the box on any standard PC or laptop with Windows, Linux, or Mac.
Genome Analysis Module
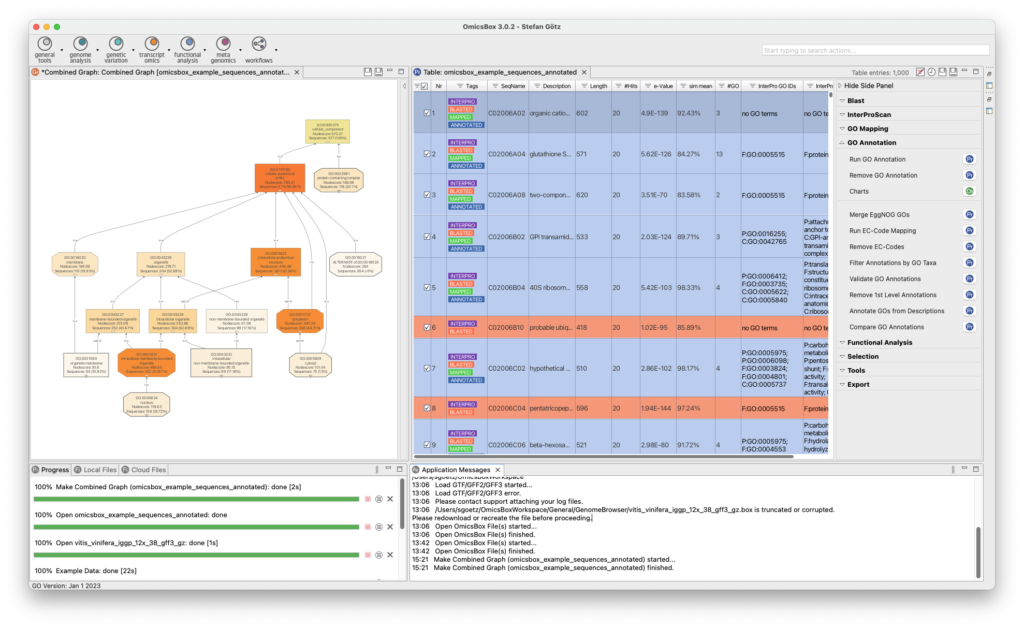
The Genome Analysis Module is an efficient and user-friendly toolset to characterizes and analyzes newly sequenced genomes. Quality control is performed using FastQC and Trimmomatic, followed by the assembly of whole genome sequences using popular algorithms such as ABySS, SPAdes, and Flye. BWA and Bowtie2 are used for read alignments, and tools for repeat identification and masking prior to gene prediction are also available. OmicsBox enables visualization of annotations through tracks that combine genome sequences, alignments, intron-exon structure, and variant data.
- Quality Control And Assessment
- Genome Mapping and Assembly
- Repeat Masking and Gene Finding
- Genome Curation Tools
Genetic Variation Module
The Genetic Variation Module allows for variant calling, filtering, and annotation, as well as genome-wide association studies to associate genetic variations with a particular trait or disease. The module offers two analysis strategies for variant calling and filtering using BCFtools and FreeBayes. The resulting VCF file can be annotated via the Variant Effect Predictor from Ensembl, and guided genome-wide association studies can be executed to identify genetic variations associated with a particular trait. This tool combination outperforms alternatives in recent review studies.
- Fast Variant Calling and Filtering
- Supports GBS and WGS data
- Custom Variant Annotation
- Genome Wide Association Studies
Transcriptomics Module
The Transcriptomics Module allows for processing RNA-Seq data from raw reads to functional analysis in a flexible and intuitive way. Quality control is followed by alignment to a reference genome using STAR or BWA or de-novo transcriptome assembly using Trinity. Coding regions can be predicted, and expression can be quantified using HTSeq or RSEM. Statistical packages like NOISeq, edgeR, or maSigPro detect differentially expressed genes. Single-Cell RNA-Seq tools identify cell groups and lineage trajectories. Enrichment analysis is integrated to identify over- and underrepresented biological functions easily.
- De-Novo Assembly and Alignment
- Differential Expression Analysis
- Long Read Transcriptomics
- Single-Cell RNA-Seq
Functional Analysis Module
The Functional Analysis Module provides biological context as an analysis option. The Blast2GO methodology allows to flexibly assigns reliable functional functional labels to novel sequences through fast alignment, domain, and orthology information generated by cloud-based Blast, InterPro, and EggNog searches. Furthermore, enrichment approaches such as Fisher’s Exact Test and GSEA identify over- and underrepresented biological functions. The Combined Pathway Analysis identifies Reactome and KEGG pathways for any sequence set, allowing pathway enrichment calculation and providing rich visualizations for insights.
- Basic Sequence Analysis
- Blast2GO Functional Annotation
- Enrichment Analysis
- Pathway Analysis with KEGG
Metagenomics Module
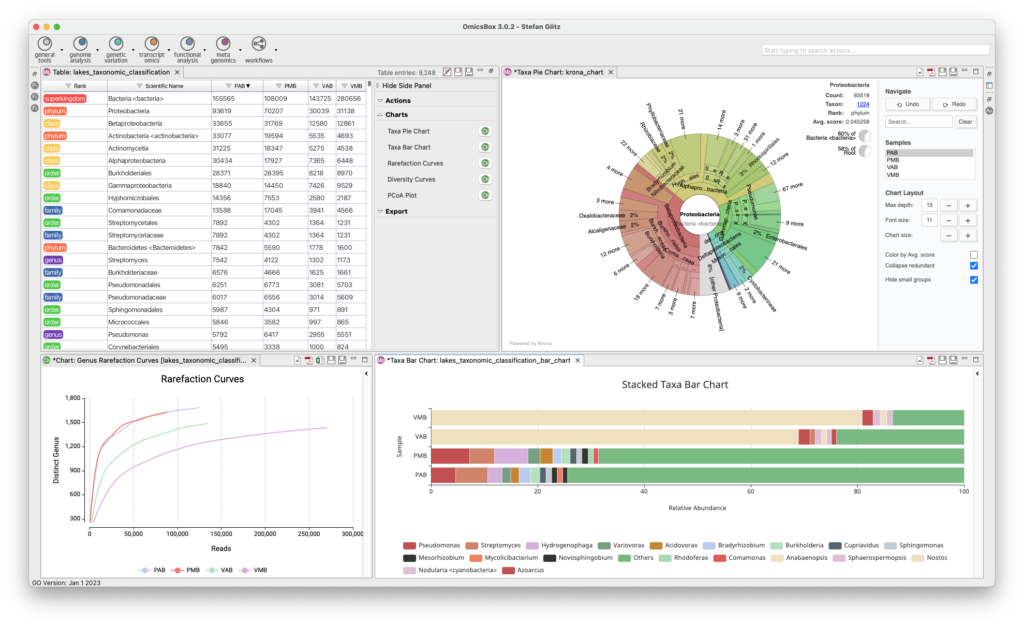
The Metagenomics Module enables the analysis of microbiome data, including assembly, annotation, and classification of metagenomic data. Kraken identifies microorganisms down to the strain level, and MetaSPAdes and MEGAHIT assemble large datasets easily and quickly in the cloud. FragGeneScan and Prodigal identify and extract possible genes and proteins, and functional annotation with various databases is available. Statistical tests can identify differential abundance of taxonomies and biological functions between samples, with rich visualizations to aid in interpretation.
- Metagenomic Assembly
- Taxonomic Classification
- Functional Annotation and Analysis
- Comparative Analysis
OmicsBox Base Platform
Manage Projects and Files, Access to Cloud Computing, Design, Run and Save Workflows, Visualize Data, Genome Browser, Filter and Sort Large Tables, General Bioinformatics Tools, and More.
- Workflows
- Genome Browser
- FastQC Quality Control Tools
- Generic Charts (Bar, Pie, etc.) and Venn Diagrams
- Access to File Manager, Support Mail, Cloud Usage
- Load, View and Export all supported file types (.box, .b2g, .fasta, . fastq, .gff, .vcf., .bam, etc.)
- Barcode Demultiplexing with Cutadapt
Over 500 institutions worldwide trust OmicsBox as their bioinformatics software