After running Gene Finding within OmicsBox/Blast2GO I ended up with a GFF containing the genome coordinates and also an OmicsBox/Blast2GO project.
With the project, I can run the full functional annotation steps Blast, InterProScan, Mapping and Annotation using the Functional Analysis Module.
Now it would be nice if I could export a GFF file retrieved from Gene Finding containing the GOs from the annotated project.
This is possible with OmicsBox/Blast2GO PRO.
To export the GFF with the GO terms first, make sure you have both GFF and the OmicsBox/Blast2GO projects saved as .box/.b2g and do the following:
- Open the Gene Finder GFF output.
- Go to File > Export > Export GFF.
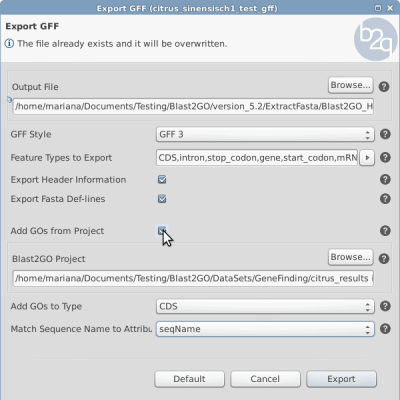
- The export parameters window will open (Figure 1).
- Select the output folder where to save the .gff file
- Select the type GFF3 and GFF2/GTF
- Features to Export (e.g. gene, CDS, mRNA, exon)
- Choose to export header, Fasta
- Add GOs from Project – this is the option that has to be enabled to add the GOs from the project to the GFF file.
- Blast2GO Project – provide the saved project that contains the functional annotation steps
- Add GOs to Type – which feature type will have the GOs (e.g. CDS)
- Match Sequence Name to Attribute – the attribute column from the GFF jas to match the Sequence Name column of the project.
- Click Export – the GFF file will be generated containing the GO ids from the Project.
With the above-mentioned instructions and these files, you will be able to export a GFF file containing the GO terms from the Blast2GO project.
Project:citrus_sinensisch1_project.b2g
GFF:citrus_sinensisch1_gff.b2g
Exported file:citrus_sinensisch1.gff