OmicsBox/Blast2GO offers two different features to retrieve the gene/protein sequences as well as the corresponding annotation from a list of identifiers within Blast2GO PRO.
Both features can be found under File > Load > Load Annotations. The expected input file is a text file with the identifiers in a single column without a header.
Use cases:
1 – Annotate the list of interest
Here I wanted to functionally annotate the list of interest without going through the annotation pipeline (blast, mapping, annotation, InterProScan).
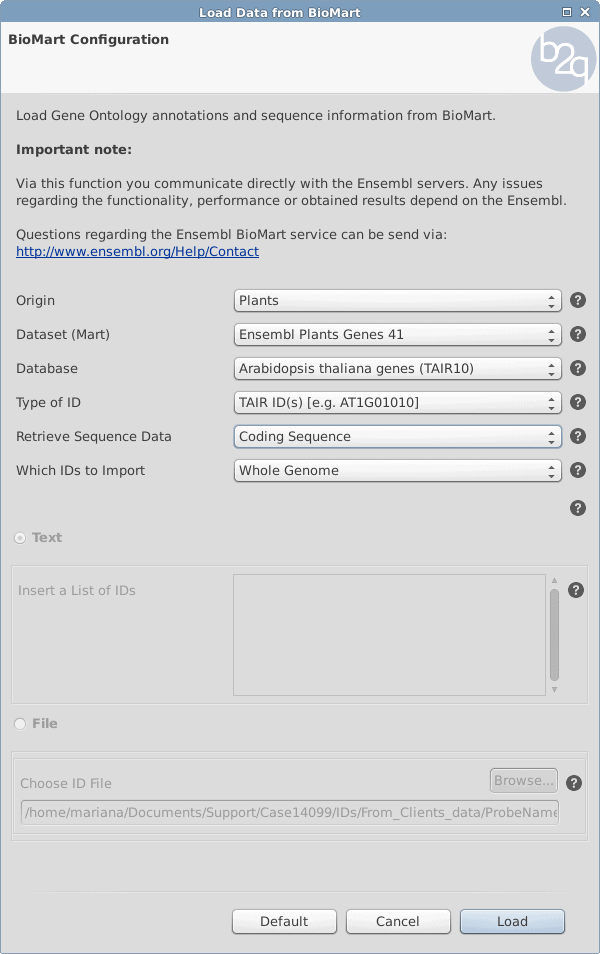
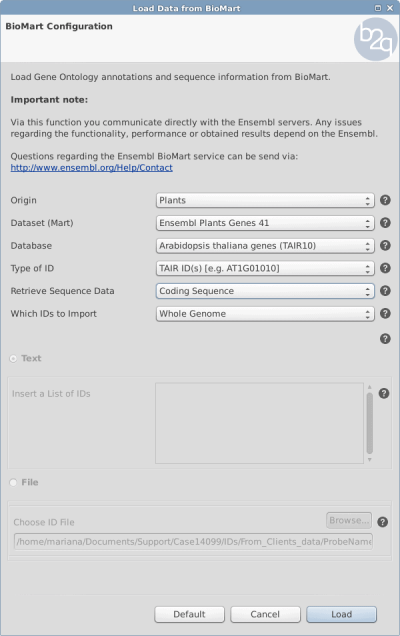
One option is to retrieve the annotations directly from BioMart. With this tool, there is the need to know the Mart, database and the type of identifiers one has.
It is not only possible to retrieve the annotations (GO terms) as well as the protein or nucleotide sequences itself.
The other one is the Load Sequence Data/ Annotation (online), which retrieves the information directly from the NCBI.
The text file, in this case, should have two columns separated by a tab, where the first column should be the identifiers (locus, proteins) and the other the taxonomy identifier.
Table 1: Example of the text files for both scenarios. A – BioMart using the Agilent ProbeNames; B – Load Sequence Data/ Annotation (online) with taxa id.
| A – Identifiers for BioMart | B – Identifiers with taxa id in tab separated | |
| A_96_P262712 | AT1G15520 | 3702 |
| A_96_P262712 | AT1G18900 | 3702 |
| A_96_P262712 | AT5G14970 | 3702 |
2 – Load annotated sequences to run functional enrichment analysis to use as the reference set
While doing a differential expression analysis, I end up with a list of interesting genes and I would like to proceed with functional enrichment analysis.
I came across a problem where I did not have an annotated project with Blast2GO that match the list of interest.
To run the functional enrichment analysis within Blast2GO an annotated project is needed.
It would possible to run the annotation pipeline (blast, mapping and annotation) of my whole genome, however, it can be very time-consuming.
In this particular case, the best would be to download the already annotated whole genome.
It is possible to do this with Blast2GO by retrieving the whole genome via BioMart in Load Annotations from BioMart (Online) or by providing a list of gene identifiers.
Using the parameters in Figure 1 a new Blast2GO Project will be created with the gene identifiers of Arabidopsis Thaliana (e.g. AT1G03850) and the corresponding annotation (GO term).
Figure 1: Retrieving the sequence whole annotated genome of Arabidopsis Thaliana with the genes identifiers
Note: If the sequences itself are not requested under the Retrieve Sequence Data parameter, then
On the other hand, it is also possible to download a list of identifiers of the species of interest from NCBI and proceed with the retrieval of the annotation using the Load Sequence Data/ Annotation (online).
In the following example, the protein identifiers from Bacterium Vibrio Vulnificus will be downloaded.
-
- Go to https://www.ncbi.nlm.nih.gov/protein/?term=bacterium%20Vibrio%20vulnificus
- Under Send to > File > Accession List the protein identifiers can be downloaded.
- This list can now be used in Blast2GO to retrieve the sequences and the annotation within Blast2GO PRO under File > Load > Load Annotations > Load Sequence Data/ Annotation (online).
Finally, I ended up with an annotated Blast2GO project that can now be used to run the functional enrichment analysis.
Note: The identifiers from the annotation project and the list of interest have to match in order to run the functional enrichment analysis. For further details on Fisher Exact Test and GSEA see the online user manual and video tutorials (How to use BioMart and GO-Slim).