Multilocus Sequence Typing MLST Analysis Tool
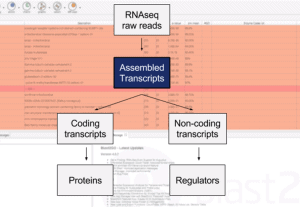
The Multilocus Sequence Typing Analysis Tool of OmicsBox (MLST) is a nucleotide sequence-based approach of characterizing isolates of bacterial species using the sequences of internal fragments of seven housekeeping genes. This video shows step-by-step how to analyze bacterial sequences with the MLST App in OmicsBox. As input, users can provide either DNA-Seq reads in FASTQ or contigs/scaffolds in FASTA format.