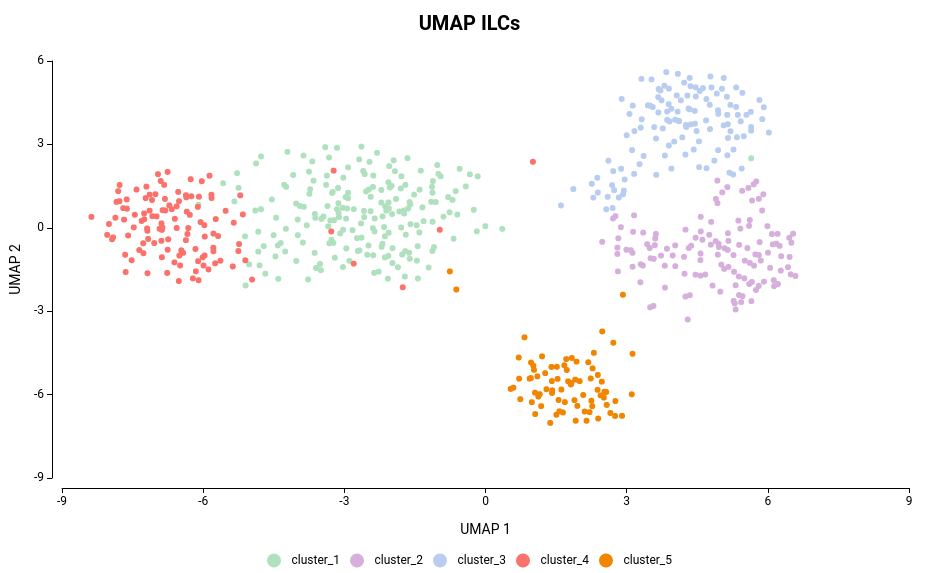
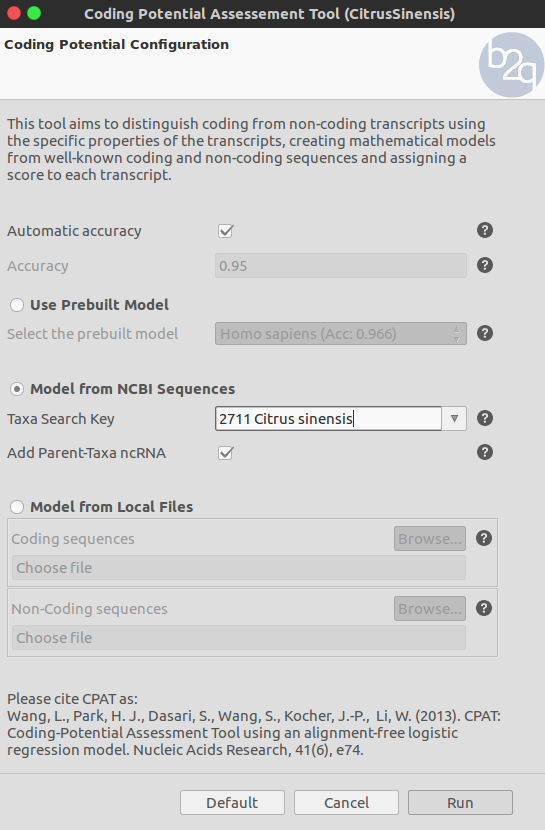
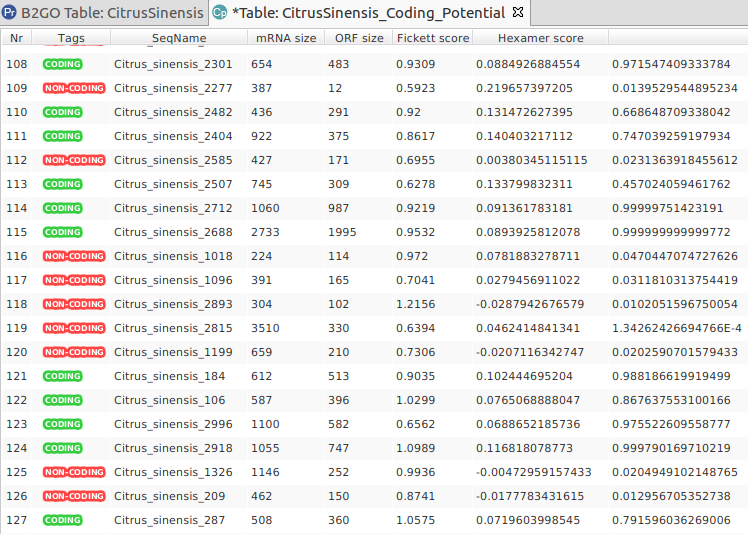
Extend the functionalities of OmicsBox
OmicsBox Apps and our Developer SDK can be used to extend the functionalities of OmicsBox. With OmicsBox Apps you are able to:
- Share your own bioinformatics tools or visualizations with the BioBam Community.
- Create a tool by wrapping a popular command line program to ease your or your colleagues’ research.
- Connect your software/system with a OmicsBox App that integrates OmicsBox into your existing environment.
Interested in making your own App?
You are more than welcome! Please write us an email to dev@biobam.com and we will provide you with a login to our developer zone and a developer licence key for OmicsBox. Looking forward to getting in touch with you.
Access to the Developer Zone allows you to use all the resources needed to develop OmicsBox Apps.
App Directory
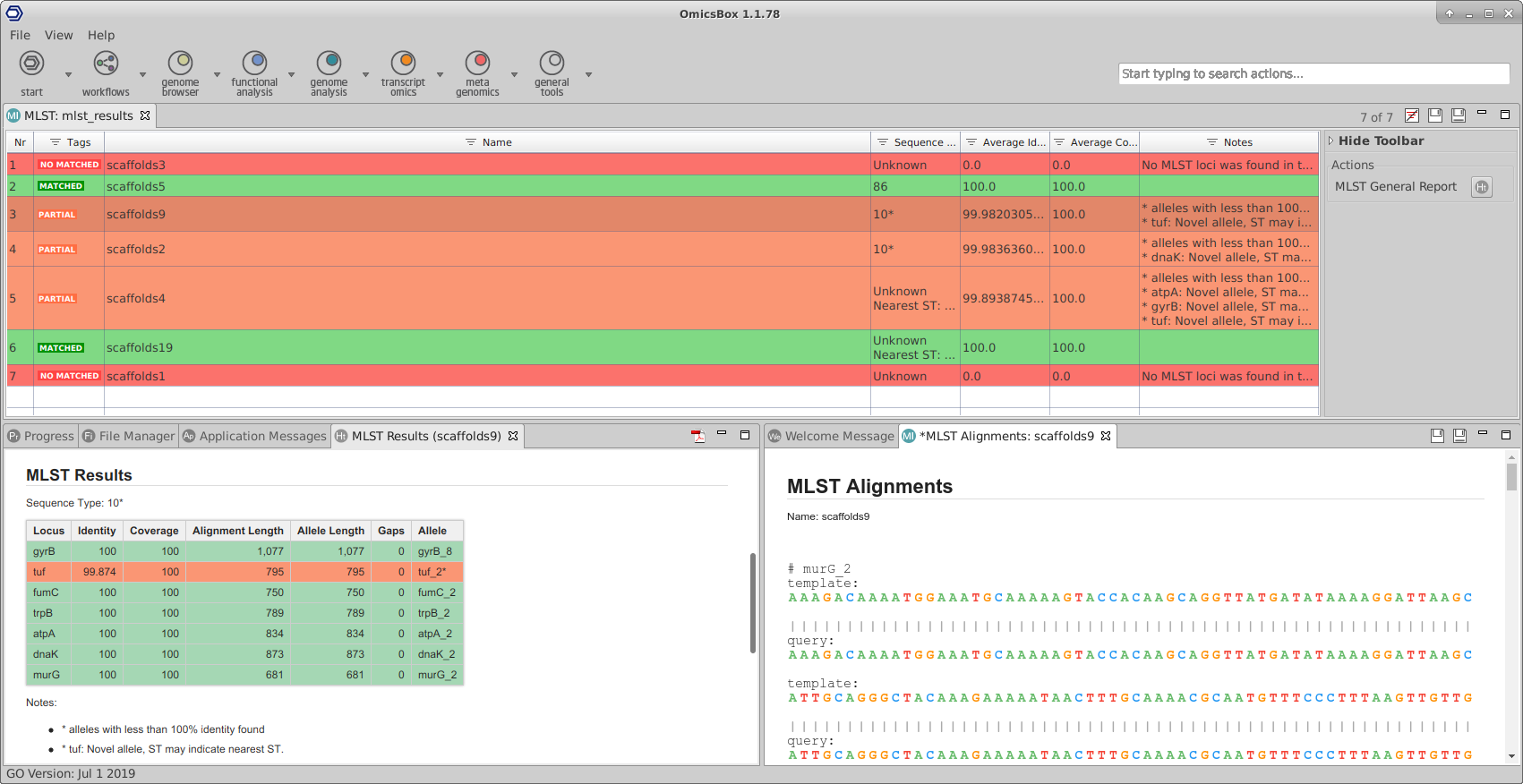
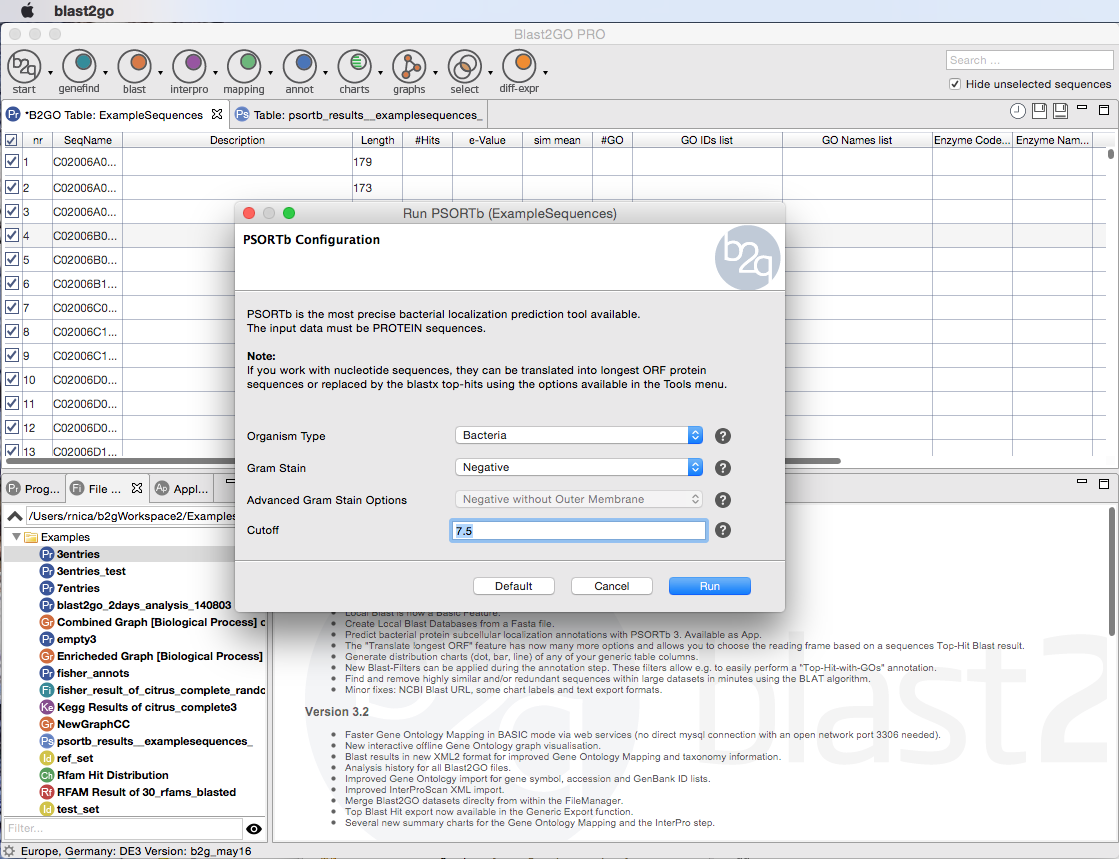
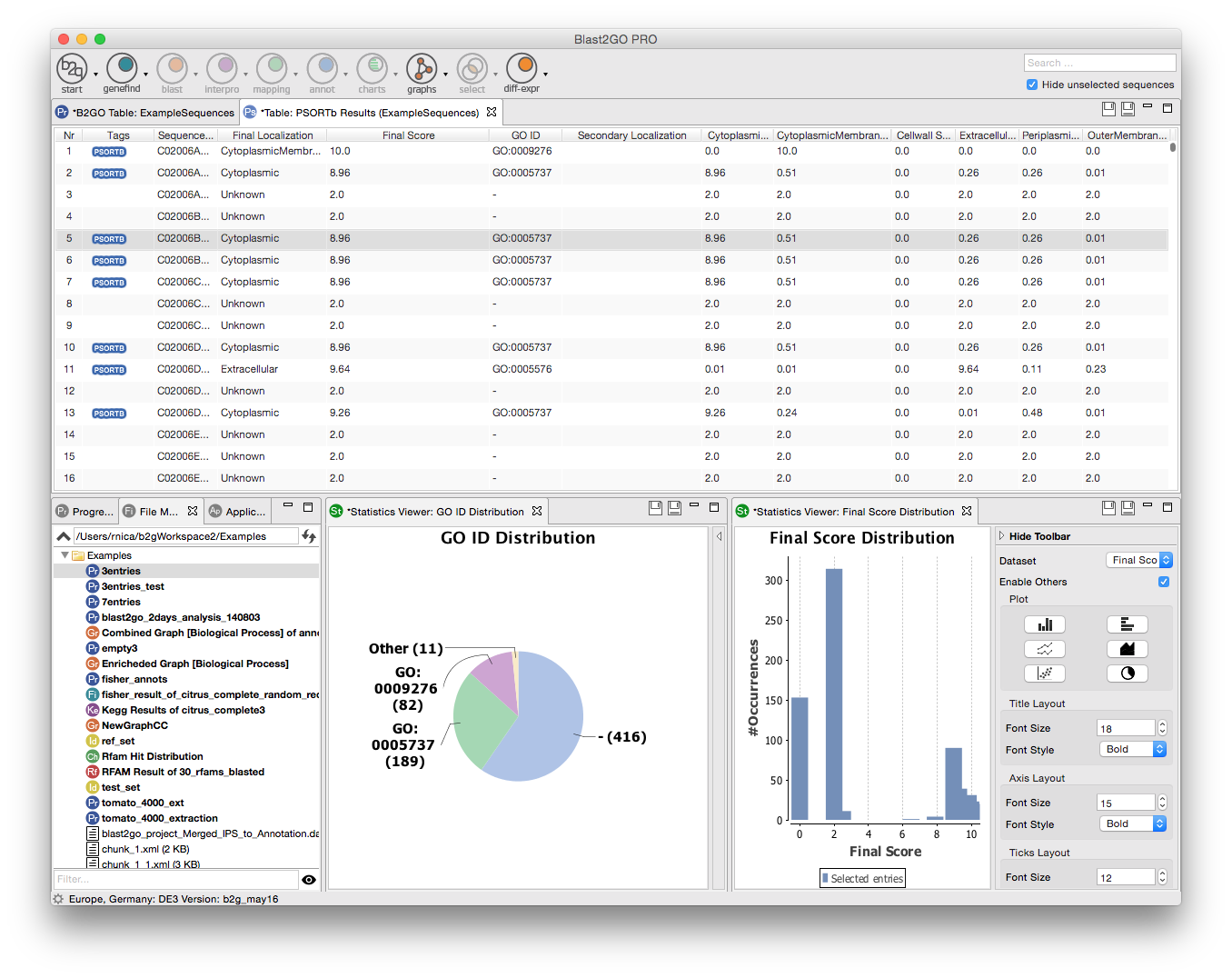
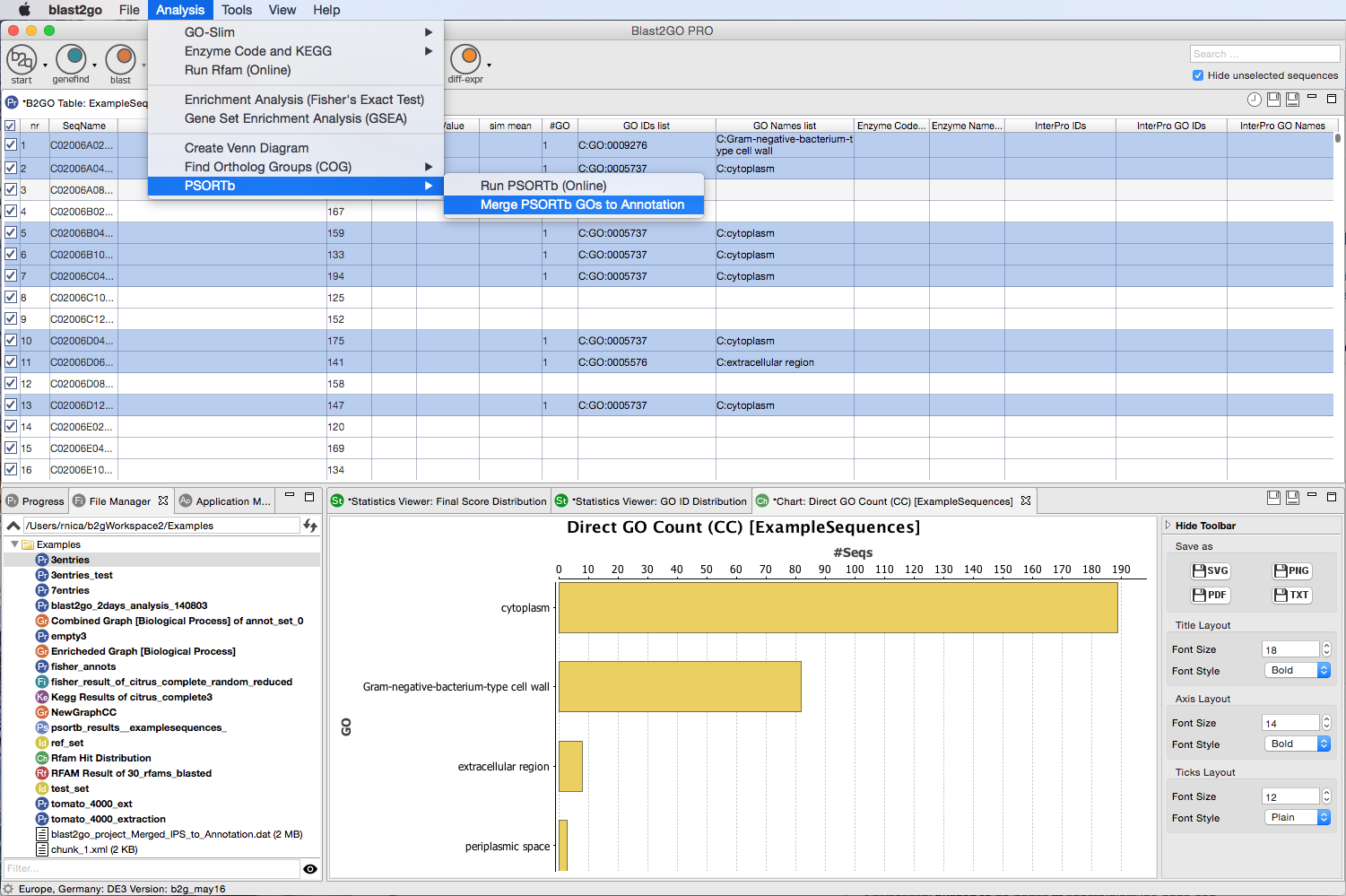
Find here a list of already available tools developed by the BioBam Community and us shortly.